Osprey
Open-source MATLAB software for processing, modeling, and quantification of magnetic resonance spectroscopy data.

Download the latest Osprey release
Read the Osprey documentation
Visit the Osprey support forum at the MRSHub
One of the challenges of magnetic resonance spectroscopy research is the complexity and abundance of analysis methods. Most researchers have created their own analysis pipelines and scripts to transform raw data into metabolite concentrations, often using code that has grown somewhat organically over the course of many years. Perhaps it should not be surprising that the results that different researchers come up with do not typically agree very well.
We actively develop Osprey, our all-in-one software environment for state-of-the art processing and quantitative analysis of in-vivo magnetic resonance spectroscopy (MRS) data. Osprey covers all steps of modern MRS data analysis, in compliance with 2020 expert consensus recommendations and adhering to minimum reporting standards.
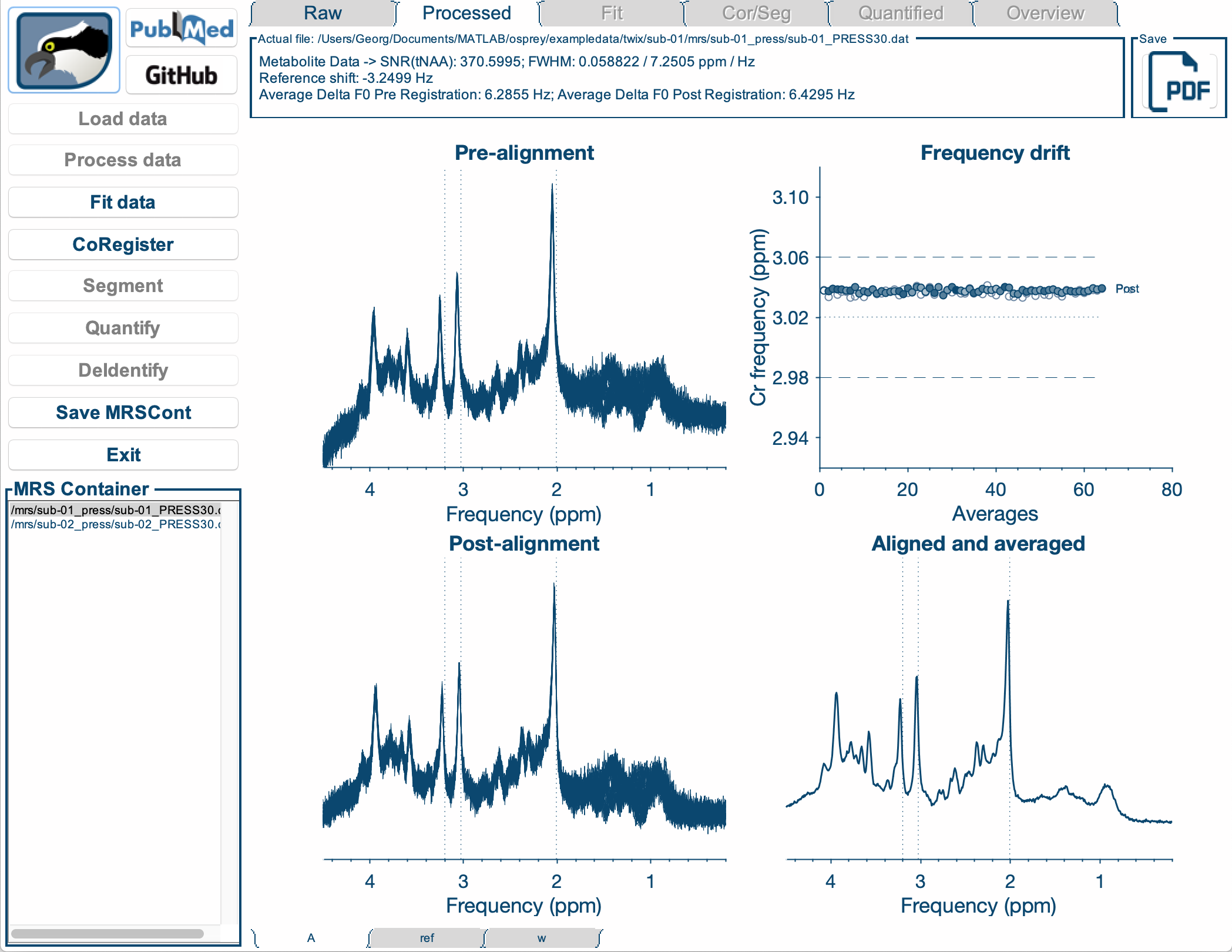
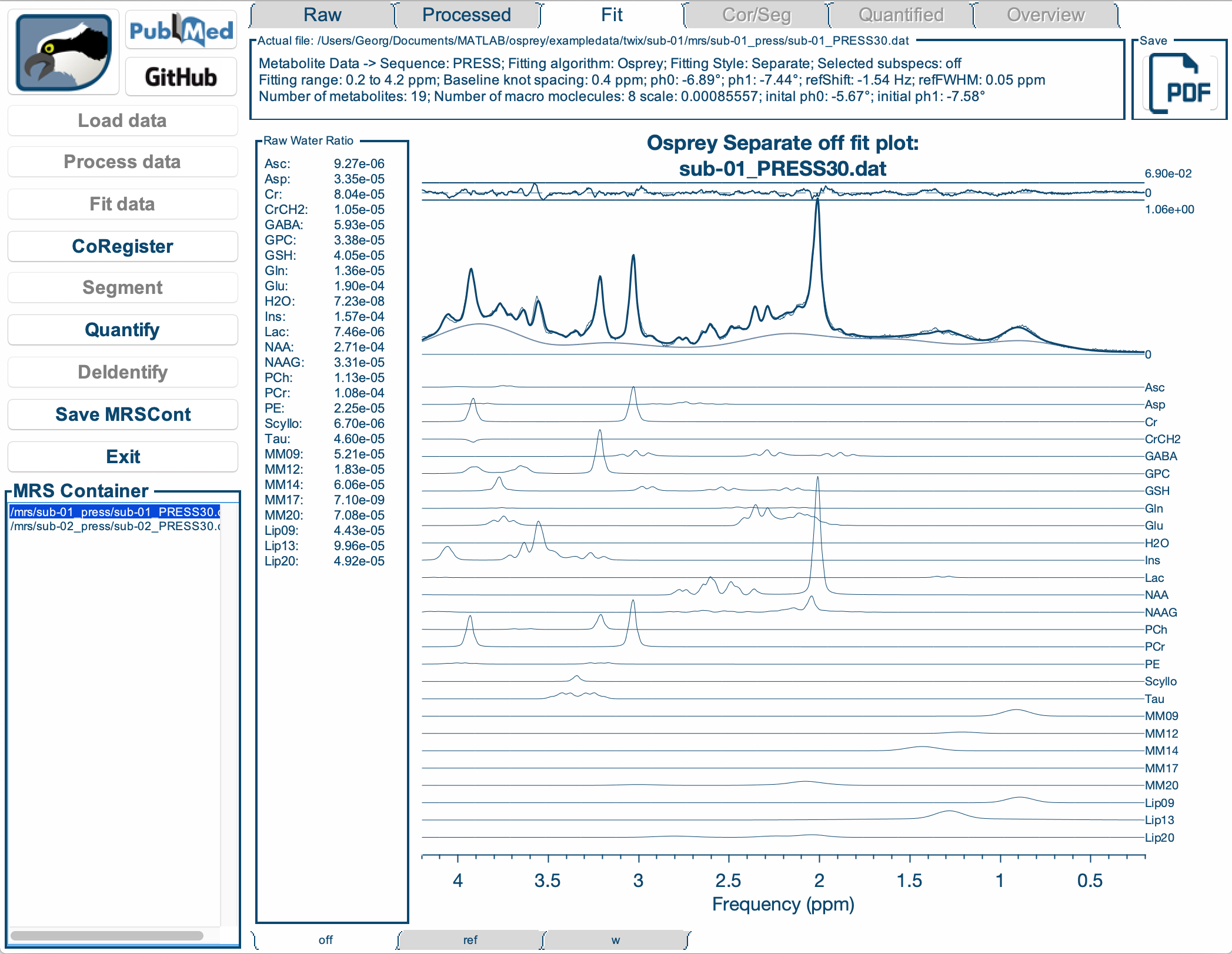
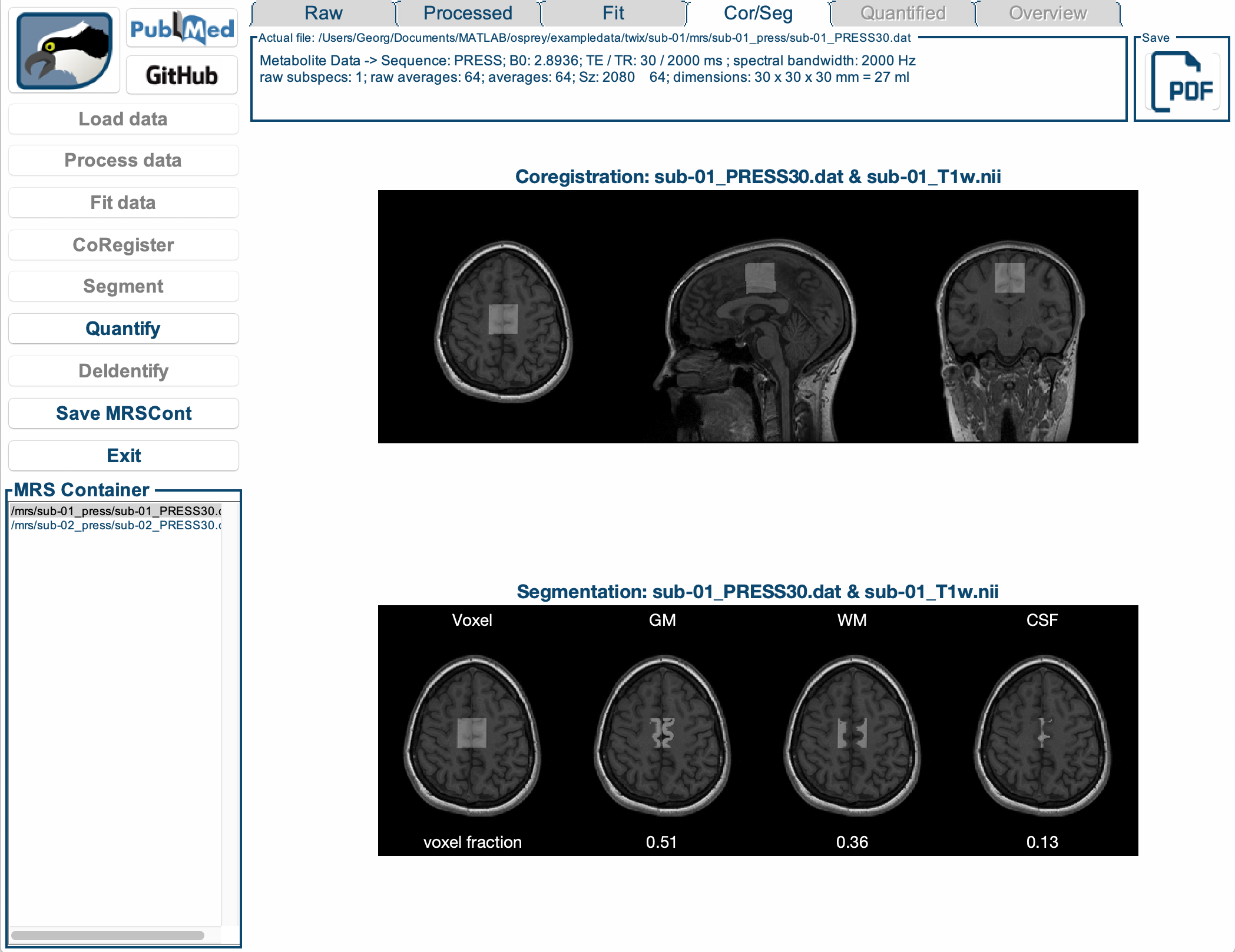
Our research is equally focused on the development and the application of advanced methodology. We created Osprey to provide a tool that can be applied out of the box by non-expert MRS users to analyze the most common MRS acquisition techniques. At the same time, the workflow is designed in a modular way to allow Osprey to easily expand and adapt to new acquisition and analysis methods.
Osprey will always be entirely free and open-source. You can download the latest release from the Osprey GitHub repository.

Download the latest Osprey release
Read the Osprey documentation
Visit the Osprey support forum at the MRSHub





